In August of 2007, the Food and Drug Administration (FDA) approved the first chemokine (C-C motif) receptor 5 (CCR5) inhibitor, maraviroc, for treatment-experienced patients infected with R5-using virus. In an effort to better educate physicians and patients about these new drugs, this article will review the available clinical data on CCR5 antagonists and discuss implications for clinical practice. Chemokine receptor antagonists are the first antiretrovirals to bind a cellular protein of the host and, as such, engender unique safety concerns. We will review the biology of HIV chemokine usage and entry into cells that first suggested the attractiveness of CCR5 blockade as a therapeutic target. We will discuss the 2 primary means of CCR5 antagonism—small molecule inhibition and monoclonal antibody steric blockade recognizing that, in the near future, only the small molecule antagonist maraviroc is FDA approved for clinical use. Based on the available clinical evidence, we provide a preliminary framework to guide the rational use of these agents in HIV-infected patients.
| Entry of HIV | Top of page |
The entry of HIV shares common mechanisms with other enveloped viruses.1 The HIV-1 surface glycoproteins gp120 and gp41 mediate viral binding to and membrane fusion with host target cells. These 2 proteins are products of the HIV env gene. The env gene product is first translated as gp160 and then intracellularly cleaved into gp120 and gp41.2 Both gp120 and gp41 traffic to the viral membrane and are the only 2 viral proteins known to protrude from its surface. On the viral membrane, there is a noncovalent association of 1 molecule of gp120 with 1 molecule of gp41.3 Three of these units aggregate on the membrane surface to form the gp120/gp41 heterotrimer (Figure 1A).4-6
Figure 1. HIV Entry
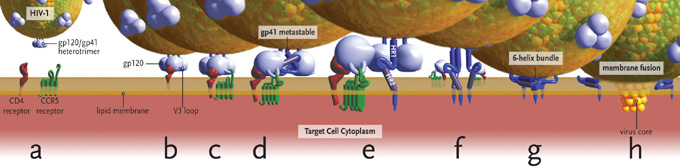
(a) Entry requires CD4, envelope glycoproteins assembled as a heterotrimer, and a coreceptor – in this case CCR5.(b) Binding of CD4 to gp120 exposes a flexible V3 loop.(c-d) The V3 loop interacts with both the N-terminus and 2nd extracellular loop (ECL2) of CCR5, leading to the release of gp41 by a currently unknown mechanism.(e) gp41 inserts into the target cell membrane.(f-g) gp41 rearranges and brings the HR1 and HR2 domains together to form a 6-helix bundle.(h) Viral and cellular membranes approximate, leading to fusion.
Images and virus models created by Louis E. Henderson, PhD
The association of gp120 with gp41 in the trimer traps gp41 in a conformationally metastable state, the energy from which can later be exploited to accelerate the rate of fusion.7 Binding of gp120 to its primary receptor on the cell surface, CD4, is the first step in membrane fusion (Figure 1B). The CD4 binding site is not completely formed in unliganded gp120, but is stabilized and fixed by the approach of CD4.8 After CD4 binding, sequential binding of the coreceptor is typically the next step in the fusion process (Figure 1C). It should be noted, however, that some interactions of CD4 with gp120 lead to inappropriate shedding of gp120 and inactivation of the fusion reaction. For fusion to proceed, gp120 must next bind its coreceptor, either CCR5 or CXCR4. The interaction of gp120 with CD4 leads to a conformational change that results in formation of the coreceptor binding site on gp120.9 Coreceptor recognition is determined by several structural elements of gp120 that include the V1-V2 region, the bridging sheet (an antiparallel, 4-stranded β sheet that connects the inner and outer domains of gp120) and, most importantly, the V3 loop.9-12 The V1-V2 stem can influence coreceptor usage through its peptide composition as well as by the degree of N-linked glycosylation.13-14 Little bridging sheet sequence variation exists in human and primate viruses and likely represents a common determinant for recognition of either coreceptor.15 The V3 loop, by contrast, is highly variable and is the principal determinant of coreceptor specificity.16-20
According to current models of HIV-1 entry, sequential binding of gp120 to CD4 and coreceptor leads to the release of gp41 from its metastable conformation (Figure 1D). The hydrophobic N-terminus, or fusion domain, of the gp41 ectodomain, now free, inserts into the target membrane (Figure 1E).7,21,22 Two trimeric coiled coil structures in gp41, termed heptad repeat 1 (HR1) and heptad repeat 2 (HR2), rearrange to form a 6-helix bundle (Figure 1F) that leads to the approximation of the 2 membranes (Figure 1G) and eventual fusion (Figure 1H).1 Some evidence does suggest that the formation of the HR1 coiled coil results from the initial binding of CD4 to gp120, before coreceptor binding occurs.23
| The HIV Coreceptor CCR5 | Top of page |
Both CCR5 and CXCR4 are chemokine receptors that belong to the large family of 7-transmembrane segment G-protein –coupled receptors. These chemokine abbreviations signify whether the first 2 cysteine (protein abbreviation C) residues of each protein are adjacent (CC) or separated by a variable amino acid (CXC), with R abbreviating receptor. During natural HIV infection, CCR5 is the primary coreceptor used. CCR5 is expressed on memory T cells, activated T cells, gastric-associated lymphoid tissue (GALT) and macrophages, and it demonstrates a greater affinity for soluble gp120 (Kd<10nM) than does CXCR4 (Kd=200-500 nM).24,25 The tyrosine sulfated N-terminus of CCR5 is the essential determinant of binding to gp120.26,27 Although not conclusively proven, the current model of gp120-CCR5 binding involves 2 critical steps: an initial interaction between sulfated tyrosines in the CCR5 N-terminus and gp120, followed by a second interaction between gp120 and the CCR5 transmembrane domains. Studies implicate both the extracellular loop 2 (ECL2) of CCR5 and/or an area at the transmembrane 4/ECL2 junction as important to this second interaction and subsequent HIV infection.27-29
| Coreceptor Usage and the V3 Loop | Top of page |
Figure 2. The V3 Loop of HIV gp120.
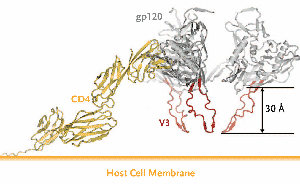
Upon CD4 binding to gp120, the V3 loop is exposed and extends towards the target cell membrane. This loop acts as a molecular “hook” that interacts with either CCR5 or CXCR4. The amino acid composition of the V3 loop is the main, but not the only, determinant of which coreceptor—CCR5 or CXCR4—a given HIV uses for entry.
From Huang C, Tang M, Zhang M, et al. Structure of a V3-Containing HIV-1 gp120 Core. Science. 2005;310:1025-1028. Reprinted with permission from American Society for the Advancement of Science (AAAS).
The V3 loop of gp120 (Figure 2) is composed of approximately 35 amino acids, and single substitutions in this region can change viral coreceptor usage.30 Although the V3 loop can be subdivided into 2 domains—the V3 stem and the V3 crown—it is the V3 crown that determines viral coreceptor tropism.31 The presence of basic amino acids within the V3 loop at positions 11 and 25 is a useful first approximation to distinguish a virus using CXCR4 for entry (X4 virus) from a virus using CCR5 (R5 virus).19,32 A coreceptor prediction algorithm solely based on amino acid composition at these 2 positions (the so-called 11/25 method) demonstrates a specificity of 90% and a positive predictive value (PPV) of 62% for detecting CXCR4 usage.33 Other more complex bioinformatics methods, such as the position-specific scoring matrices (PSSMs), analyze the entire V3 sequence to predict coreceptor usage.34 These models do not account for sequences outside of the V3 loop, for example, the V2 or C4 (contains the bridging sheet) regions, that may also influence coreceptor tropism.35 Some viruses can use both CXCR4 and CCR5 for viral entry and are referred to as R5/X4, or dual-tropic viruses.36,37
Over 90% of primary HIV infection occurs with R5 virus and appears to represent a pure R5 population as determined by coreceptor tropism assays.38-42 Coreceptor usage, however, is a dynamic process. Within 5 years of infection, approximately 50% of patients with HIV have detectable populations of syncytium-inducing (SI) viruses—a phenotype linked to CXCR4 usage.43 Although the mechanisms of this phenotypic switch are not clearly understood, the clinical consequences are well described. The appearance of SI virus is associated with a faster rate of CD4+ T-cell loss, rapid disease progression, and an increased rate of development of AIDS and death.44-47 This may be due in part to the increased cytopathic effect of X4-tropic virus and the increased T-cell target population accessible to this virus.48 Assays for syncytium induction, however, are not routinely performed on clinical samples. For this reason, the use of the 11/25 method mentioned above was investigated as a simpler way to predict both phenotype and clinical course. In a study of patients receiving triple antiretroviral therapy, the 11/25 genotype of the V3 loop alone was an independent predictor of both poor immunologic response and more rapid mortality.49 Emergence of X4 virus does not occur in all patients who die of AIDS. In some patients, progression of their clinical course is associated with increased virulence of pre-existing R5 virus.50
Recently, an analysis of existing data sets from the AIDS Clinical Trials Group 5211 (ACTG 5211) and the T-20 Versus Optimized Regimen Only (TORO) trials using the Trofile™ tropism assay (see below) interestingly demonstrates that patients with only X4 virus have CD4 counts similar to patients with only R5 virus and have lower baseline HIV RNA levels than either R5 or dual-mixed viral populations.51,52 That is, the more rapid disease progression seen with the X4 virus may, in fact, be due to infections with dual-tropic or mixed R5X4 viruses, intimating that the R5 component may have been missed with prior assays geared to detect the syncytium-inducing phenotype alone. As such, infection with pure X4 virus would be less of a concern than infection with dual-tropic or mixed viral populations.
| Determining Coreceptor Usage in Clinical Samples | Top of page |
Since CCR5 antagonists have anti-HIV activity only against R5-using viruses, all patients being considered for CCR5 antagonist therapy will need a blood test to determine their virus’ coreceptor usage. The drug label for maraviroc states it should be used in treatment-experienced patients with documented R5 virus (only). This test is required because the use of an R5 antagonist as part of a triple regimen in patients with pure X4 virus exposes the patient to suboptimal therapy with all the concomitant associated risks of virologic failure. Further, the use of an R5 antagonist in patients with R5/X4 dual/mixed-tropic virus may select for the emergence of X4 virus. Clearly, knowing the patient’s viral coreceptor usage, or tropism, is critical to the appropriate use of these drugs in day-to-day clinical practice.
In the United States, the Trofile™ HIV coreceptor tropism assay (Monogram Biosciences) will be commercially available to determine viral tropism and should have a turnaround time of between 2 and 3 weeks. Before discussing the 2 tests, one point deserves clarification: viral tropism technically refers to the types of cells (eg, T cells, macrophages) that a virus can infect, while coreceptor usage defines a virus as either R5 or X4.53 However, since “tropism” is so often used interchangeably in the literature with coreceptor usage, we will follow the now common usage of tropism and will refer to viruses as either R5 or X4 tropic.
| The Trofile™ HIV Coreceptor Tropism Assay | Top of page |
The Trofile™ assay, developed and marketed by Monogram Biosciences, determines tropism by isolating and cloning full-length, patient-derived viral env from patient blood samples into a proprietary vector system.54 The cost of this test is approximately $1,960. As with resistance testing, viral loads above 1,000 copies per mL are needed to most accurately perform this test. Two separate vectors are used: one contains the cloned patient envelope genes and the other contains the remainder of the HIV genome. The HIV genomic vector contains an intentional self-inactivating deletion in the long terminal repeat (LTR) region that effectively prevents more than 1 round of viral replication. Thus, the Trofile™ assay is an example of a single-cycle assay.
Neither vector alone can produce virions, so both the envelope expression vector and the HIV genomic vector are cotransfected (ie, inserted) into the same laboratory cell line for the purposes of generating infectious viruses. These viruses are harvested and then used to infect a malignant glioma cell line (U87 cells) that have been engineered to express CD4 and either CCR5 or CXCR4 on their surface. An enzyme (luciferase) expressed from the genomic vector generates light when the reporter cell line becomes infected by virus. Any light produced can then be measured and quantified. So, for a given patient sample, the amount of light generated in either the CCR5 or CXCR4-expressing cells will reveal whether the virus uses CCR5, CXCR4, or both to enter cells. The specificity of the assay is improved by subsequently seeing how much of the generated light can be blocked by the addition of an R5- or X4-specific antagonist into the cell cultures. Test results supplied to the clinician will indicate whether a virus is R5-tropic, X4-tropic, or dual/mixed. The term dual/mixed refers to the fact that the Trofile™ assay cannot distinguish between the presence of one virus that uses either receptor for viral entry (dual-tropic) or mixed viral populations in the same patient sample that uses either CCR5 or CXCR4. For clinical purposes, this distinction is not as important as knowing whether the sample contains any X4-using virus.
| The Tropism Recombinant Test (Phenoscript™ Assay) | Top of page |
The Phenoscript™ assay (Eurofins VIRalliance Inc.) is another recombinant phenotypic assay that will be commercially available for the determination of viral coreceptor usage, although it has not been cross-validated with the Monogram Trofile™ assay.55 Virus is isolated from patient blood and only a portion of the HIV envelope gene is amplified. This 900bp piece contains V1-V3 (the 1st, 2nd, and 3rd variable loops), which contains the major determinants of coreceptor usage. In this assay, only 1 vector is used that contains the HIV genome with a 900bp deletion in the env gene. The patient-derived V1-V3 envelope piece is recombined with the vector, corrects for the 900bp deletion, and leads to the production of infectious viruses. These viruses then infect a cell line (similar but distinct from those used in the Trofile™ assay) that has CD4 and either CCR5 or CXCR4 on its surface. With Phenoscript™, infection produces color (rather than light) that can be measured and quantified. Again, test results will report either R5-tropism, X4-tropism, or dual/mixed-tropism. Although this assay is in development, it has not been used in any of the clinical trials of CCR5 antagonists, so the interpretation of results will be more challenging (eg, “How meaningful is a low level of X4 activity?” “How effective is the assay at screening out dual/mixed virus?”).
| Analysis—Advantages and Challenges in Tropism Assays | Top of page |
Considerable technical differences between the 2 tests preclude a direct comparison of the tropism results obtained. A recent article that attempted to do just that found an 85% concordance between the 2 tests in 74 clinical isolates tested, although the absence of a gold-standard test made it impossible to determine which test, in fact, delivers the “correct” answer.55 One important limitation of both tests is the inability to reliably detect minority tropism variants that comprise less than 10% of the total viral population. Monogram has published data reporting limits of detection down to 5%, but no information is provided on the actual amount of virus used to generate those results.54 In other words, the detection of a tropism variant comprising 5% of a viral population in a patient with a viral load of 5,000 copies per mL is quite different, and considerably more difficult, than being able to detect 5% of a population in a patient with a viral load of 500,000 copies per mL.
The use of full-length envelope by the Trofile™ assay as compared with 900bp of envelope (roughly one-third) in the Phenoscript™ assay may be expected to give tropism results more representative of the actual viruses, and the use of inhibitors to improve specificity in the Trofile™ assay is a clear strength of this technique. Differences in tropism determination that may result from the use of different cell lines, variable recombination efficiency of PCR products in the Phenoscript™ assay, and improved efficiency of PCR amplification for smaller-sized amplicons (Phenoscript™ relative to Trofile™) have not yet been systematically evaluated, and their clinical relevance remains uncertain.
Tropism determination with the Trofile™ assay can be a dynamic process, with minor changes seen from week to week. Of the roughly 118 patients enrolled in ACTG 5211, a phase 2b trial evaluating the safety and efficacy of the CCR5 antagonist vicriviroc, 10% had a change from R5 to dual-mixed virus in between study screening and study inception.52 The exact reasons for this remain to be elucidated. It will be interesting to follow the literature to see whether any difference in clinical outcome in patients undergoing R5 antagonist therapy can be linked to a switch in coreceptor usage as determined by either tropism assay.
| CCR5 Antagonists | Top of page |
Coreceptor binding is essential for HIV entry. Antagonism of gp120 binding to CCR5 therefore presents an attractive target for therapeutic drug design. Earlier efforts to block the gp120 and CCR5 interaction employed chemical modifications of the known CCR5 ligands, especially RANTES (regulated on activation, normal T expressed and secreted). These compounds suffered, however, from poor oral bioavailability and the induction of intracellular signaling cascades upon CCR5 binding.25 The most promising drugs in this class, however, are nonpeptidic, orally bioavailable, small-molecule antagonists of CCR5. These small molecules are noncompetitive allosteric antagonists of CCR5 with molecular weights of 500 to 600. As allosteric inhibitors, these antagonists do not have to induce conformational changes in CCR5 per se, but may alter the binding affinity of gp120 by perturbing the equilibrium between different CCR5 conformations that are naturally present. The CCR5 antagonists have been given generic names with the suffix “-viroc,” an abbreviation for “viral receptor occupancy.”
| Aplaviroc | Top of page |
Aplaviroc was a GlaxoSmithKline compound that demonstrated antiviral activity with minimal toxicities during short-term monotherapy studies.57 Aplaviroc in various dosing regimens led to a 1.0 to 1.6 log10 reduction in viral loads during 10 days of treatment.58 In phase 2b trials, however, 4 out of roughly 300 patients developed severe hepatotoxicity that, on liver biopsy, was found to be consistent with drug-induced hepatitis. This finding led to the cessation of the trial.59 Similarly, 1 out of 26 patients participating in a phase 3 trial of aplaviroc demonstrated elevation of alanine aminotransferase to 24 times normal levels. No deaths occurred, and the hepatitis resolved with aplaviroc discontinuation. This trial was also stopped, and clinical development of aplaviroc was terminated.
| Maraviroc | Top of page |
Maraviroc is a CCR5 antagonist with potent in vitro and in vivo anti –HIV-1 activity that was recently approved by the FDA for treatment-experienced patients with R5-tropic virus. In a 10-day monotherapy trial conducted in HIV-1-infected subjects with R5 virus, administration of maraviroc at doses up to 600 mg daily resulted in a reduction of plasma HIV-1 of at least 1.6 log10.60 A shift in viral tropism, however, was noted in 2 subjects. One subject experienced transient emergence of a mixed tropic population on day 11 of therapy that was not detectable on day 40. The other developed a dual- or mixed-tropic virus by day 11 that remained detectable through day 433 of follow-up. Additional studies suggested that these viruses emerged from a pretreatment reservoir of X4 viruses that was not detected on initial tropism testing.61
The Maraviroc Versus Efavirenz Regimens as Initial Therapy (MERIT) study evaluated the activity of maraviroc versus efavirenz, both in combination with zidovudine/lamivudine (ZDV/3TC), in treatment-naive subjects with HIV-1 infection.62 Inferior efficacy compared with a standard efavirenz-based regimen led to discontinuation of the 300-mg, once-daily arm in the treatment-naive trial, but the 300-mg, twice-daily arm continued. Over 48 weeks, twice-daily maraviroc plus ZDV/3TC failed to demonstrate noninferiority to efavirenz plus ZDV/3TC in the primary endpoint of HIV-1 RNA below 50 copies per mL. One subject receiving maraviroc at a dose of 300 mg daily, in combination with ZDV/3TC, developed severe hepatotoxicity that necessitated liver transplantation.63 The subject was infected with hepatitis C virus (HCV) without detectable HCV RNA and had recently initiated other potentially hepatotoxic medications, including isoniazid and trimethoprim/sulfamethoxazole. This patient had elevated transaminases prior to receiving the first dose of maraviroc. The causal role of maraviroc in this patient’s liver failure is therefore unclear. Similar cases of severe hepatotoxicity have not been observed in ongoing or completed maraviroc studies.
Results from 2 nearly identical phase 2b/3 trials of maraviroc in antiretroviral-experienced subjects,Maraviroc Plus Optimized Therapy in Viremic Antiretroviral Treatment Experienced Patients (MOTIVATE) 1 and 2, were reported in abstract form.64,65 Enrollment criteria included HIV-1 RNA above 5,000 copies per mL, resistance to and/or at least 6 months experience with 1 or more antiretroviral from 3 classes (≥2 for protease inhibitors [PIs]), and R5 virus only detected by the Trofile™ tropism assay. These trials enrolled 601 and 475 patients, respectively, with a primary endpoint of mean change from baseline HIV-1 RNA at 24 weeks. Patients were randomized in a 1:2:2 ratio to 1 of 3 treatment arms: optimized background regimen (OBR)+placebo, OBR+maraviroc 150 mg once daily, or OBR+maraviroc 150 mg twice daily. The dose of maraviroc was adjusted for concomitant medications. Optimized background regimen therapy consisted of between 3 and 6 drugs. As would be expected for this patient population, 44% of subjects who were screened for trial eligibility had dual/mixed or X4 virus and were excluded from study participation.
In MOTIVATE 1 (USA and Canada), of the 601 patients enrolled, 585 were treated. Roughly 90% of patients were male and 80% were white. The median entry CD4 counts were 150 to 168 cells per mm3 with viral loads of 4.84 to 4.86 log10 copies per mL. Forty percent of patients received a regimen containing enfuvirtide, while approximately 70% of subjects had 2 or fewer active drugs in their OBR. Mean change in HIV-1 viral load was –1.08 log10 copies per mL for placebo, –1.82 log10 copies per mL for OBR+maraviroc QD, and –1.95 log10 copies per mL for OBR+maraviroc BID. The difference between viral load reduction with once- and twice-daily maraviroc was significantly different from placebo (P<.0001 and P=.0005, respectively), but not significantly different from each other. Similar log10 reductions in viral load were seen in subjects with viral loads greater or less than 100,000 copies per mL. Approximately 42% to 49% of subjects receiving maraviroc achieved a viral load below 50 copies compared with 25% of subjects in the placebo arm at 24 weeks. At 24 weeks, CD4 counts increased by 52 cells in the placebo arm and 107 and 111 cells per mm3 in the maraviroc once- and twice-daily arms, respectively. Increased adverse events in subjects receiving maraviroc included dizziness and upper respiratory infections. Grade 3 and 4 elevations of alanine transaminase (ALT) were more common in patients receiving maraviroc, although these data were unadjusted for duration of exposure to maraviroc. Three non-Hodgkin’s lymphomas were reported, 2 in the placebo arm and 1 in the maraviroc once-daily dosing arm.
In MOTIVATE 2 (United States, Europe, and Australia), 464 of 475 enrolled subjects received treatment with CD4 counts of 174 to 182 cells per mm3 and mean viral loads between 4.84 and 4.89 log10 copies per mL. Mean change in HIV-1 viral load at 24 weeks was –0.93 log reduction for placebo, and –1.95 and –1.97 for OBR+maraviroc once and twice daily, respectively. Viral load reductions to below 50 copies per mL were seen in 45.6% of subjects receiving maraviroc QD as compared with 40.9% receiving maraviroc BID and 20.9% receiving placebo. Mean CD4 count increases were 64 cells per mm3 in placebo, 112 cells per mm3 in OBR+maraviroc QD, and 102 cells per mm3 in OBR+maraviroc BID. For adverse events occurring in at least 10% of subjects in any group, nasopharyngitis, pyrexia, and cough were seen more often in subjects receiving maraviroc. Further, 2 cases of lymphoma were reported in patients receiving maraviroc, with none occurring in placebo subjects. This rate is consistent with historic controls and is not thought to be related to maraviroc itself. In contrast to MOTIVATE 1, no increase in liver enzyme elevations was seen in subjects taking maraviroc over those taking placebo.
A combined analysis of the 2 maraviroc trials showed, as has been seen in the Performance of TMC114/r When Evaluated in Treatment-experienced Patients with PI Resistance (POWER) and the Randomized Evaluation of Strategic Intervention in Multi-drug Resistant Patients with Tipranavir (RESIST) trials of darunavir and tipranavir, that inclusion of at least 2 active drugs in the antiretroviral regimen results in a greater percentage of subjects achieving viral loads of less than 50 copies per mL (Figure 3). Subjects receiving enfuvirtide for the first time (ie, enfuvirtide naive) in combination with maraviroc had greater rates of virologic response (HIV-1 RNA <50 copies/mL) than enfuvirtide-experienced and/or -resistant subjects. Interestingly, for patients taking at least 3 active drugs, similar proportions in the placebo and maraviroc dosing arms achieved undetectable RNA levels. In subjects experiencing virologic failure while undergoing a maraviroc-containing regimen, 53% experienced a change in viral tropism from R5 to either dual/mixed or X4 virus. In those subjects, a change in viral tropism was associated with more modest increases in CD4 cell counts (+37 and +56 cells/mm3 for maraviroc QD and BID vs +61 and +138 in subjects maintaining R5 tropism). Preliminary data indicate that maraviroc resistance is characterized by decreases in the percent maximal suppression and not with the more classic “rightward” shifts in 50% inhibitory concentration (IC50) curves. Resistance is associated with mutations in the V3 loop of gp120 (and elsewhere), but no signature mutations have yet been identified.
Figure 3. Treatment Outcome in MOTIVATE 1 and 2 Studies.
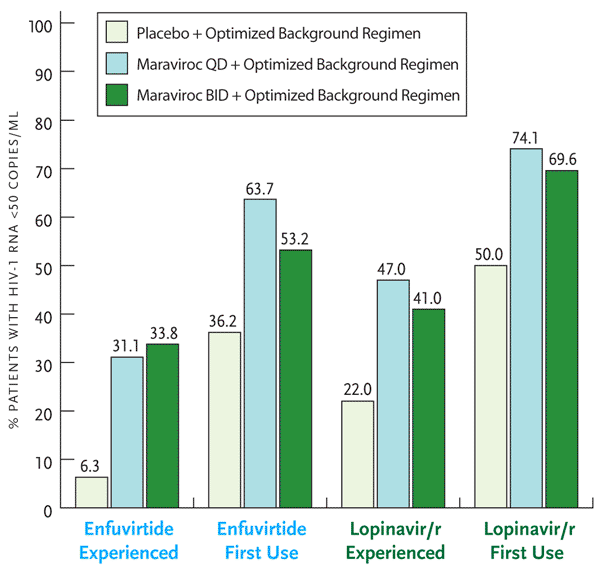
Inclusion of maraviroc and at least one other active antiretroviral, either enfuvirtide or lopinavir/ritonavir depicted here, led to a greater percentage of subjects achieving viral loads <50 copies/mL. Note that only twice–daily maraviroc is FDA approved for clinical use.
Adapted with permission from van der Ryst E, et al. Efficacy of maraviroc in combination with at least one other potent new antiretroviral drug: 24-week combined analysis of the MOTIVATE 1 and 2 studies. 4th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention. July 22-25, 2007. Sydney, Australia. Abstract WEPEB115LB.
| Vicriviroc | Top of page |
Vicriviroc, formerly known as SCH-D, is 2- to 40-fold more potent in vitro than the first-generation compound, Schering C (SCH-C). Like maraviroc, this molecule blocks signaling by the C-C chemokines at nanomolar concentrations.66 Testing in healthy volunteers or HIV-1-infected subjects has not, to date, revealed QTc prolongation or central nervous system (CNS) adverse effects. Vicriviroc is metabolized by CYP3A4 and, thus, can be boosted with ritonavir. Data from a 14-day monotherapy trial demonstrated a reduction of plasma HIV-1 RNA by approximately 1.0 to 1.5 log10 copies per mL.62,67,68 A phase 2 trial of vicriviroc in treatment-naïve HIV-1-infected subjects was discontinued because of increased rates of virologic relapse in the vicriviroc arms compared with the control efavirenz arm (all subjects also received zidovudine plus lamivudine as a fixed-dose combination (ZDV/3TC).69 Subjects randomized to the vicriviroc arms received an initial 14-day period of vicriviroc monotherapy prior to the addition of ZDV/3TC. Twenty-eight of 68 subjects (38%) receiving vicriviroc experienced virologic breakthrough. Genotypic analysis demonstrated that virus from all subjects who experienced virologic breakthrough developed the M184V mutation for 3TC resistance. This finding supports the concern that vicriviroc may have been underdosed in this study.
As mentioned, vicriviroc was recently evaluated in a phase 2b trial in antiretroviral-experienced patients by the ACTG 5211. That study compared the safety and efficacy of 3 different vicriviroc dosages in combination with an OBR to OBR alone.70 The primary endpoint was change in plasma HIV-1 RNA at day 14, with a secondary endpoint of the safety, tolerability, and HIV-1 RNA changes at 24 weeks. At 14 days and 24 weeks, mean changes in HIV-1 RNA (log10 copies/mL) were greater in the vicriviroc groups, -0.87 and -1.51 (5 mg), -1.15 and -1.86 (10 mg), and -0.92 and -1.68 (15 mg), than in the placebo group, +0.06 and –0.29 (P<.01). Grade 3/4 adverse events were similar across groups. The 48-week results were recently reported and demonstrated a sustained virologic response in subjects receiving vicriviroc (Figure 4).71
Figure 4. 8-Week Data from ACTG 5211.
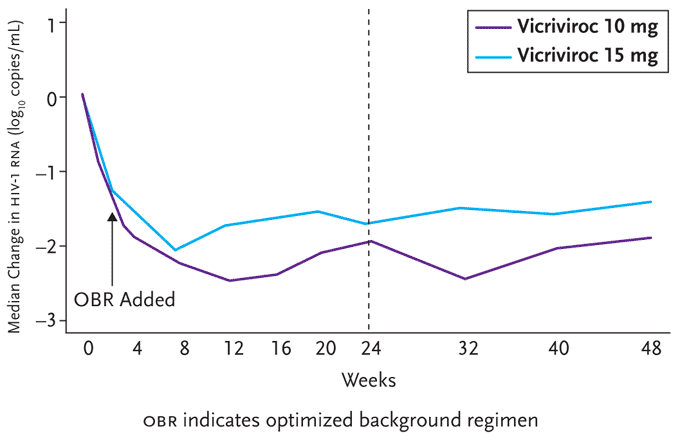
Benefits in viral load reductions previously reported after 24 weeks of follow-up were maintained at 48 weeks in both the vicriviroc 10mg and 15mg once daily arms of this trial.
Reprinted with permission from Gulick R, Su Z, Flexner C, et al. ACTG 5211: Phase II study of the safety and efficacy of vicriviroc (VCV) in HIV-infected treatment-experienced subjects: 48 week results. 4th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention. July 22-25, 2007. Sydney, Australia. Abstract TUAB102.
Virologic failure was, however, associated with a change in coreceptor usage in 35% of vicriviroc-treated subjects. Interim results raised concern when 5 cases of malignancy occurred in subjects receiving vicriviroc: 1 case of recurrent Hodgkin’s disease; a case of non-Hodgkin’s disease in a subject with previously treated Hodgkin’s; 2 de novo cases of lymphoma (1 Hodgkin’s and 1 non-Hodgkin’s); and 1 case of gastric adenocarcinoma.70 No lymphomas have been reported to date in the control arm.
Because of the small sample size (N=118) and the participants’ advanced stage of HIV disease, the significance of these results is unclear, and a causal link with vicriviroc has yet to be established. No link to increased Epstein-Barr virus (EBV) viral replication was seen in subjects receiving vicriviroc as part of this study.72 Vicriviroc has not previously been shown to be carcinogenic or mutagenic, and preclinical toxicology studies in animals showed no increased malignancy risk. To date, severe hepatitis has not been observed in clinical trials of vicriviroc.
| Other CCR5 Inhibitors | Top of page |
In a recent phase 1 study, INCB009471 (Incyte)—a novel, orally available CCR5 antagonist—was well tolerated and demonstrated a reduction of 1.7 log10 copies per mL in HIV-1 RNA levels after 14 days of monotherapy.73 Phase 2 studies are planned. In addition to small-molecule CCR5 antagonists, several monoclonal antibodies directed against CCR5 are being developed, including PRO140 (Progenics) and HGS004 (Human Genome Sciences). Given the oral bioavailability and advanced stage of clinical development of the small-molecule inhibitors, it is unclear what role CCR5 monoclonal antibodies may play in future antiretroviral therapy regimens. CCR5 monoclonal antibodies could have a distinct mechanism of action, presumably sterically hindering gp120-CCR5 binding through the much larger size of the antibody structure. Compelling clinical data are not yet available for this class of entry-inhibitor compounds.
| HIV-1 Resistance to CCR5 Inhibitors | Top of page |
The high error rate of HIV-1 reverse transcriptase and rapid turnover of the viral population facilitate the emergence of drug-resistant mutants under conditions of partial drug efficacy. In the case of CCR5 inhibitors, the drug target is a host cell protein that will not undergo mutation in response to CCR5 antagonist therapy. Viral adaptation to CCR5 inhibitors could, however, involve changes in the viral envelope protein that alters dependence on CCR5. Of greatest theoretical concern is the potential selection of CXCR4-tropic virus in response to CCR5 blockade. Whether emergence of X4 viruses in this setting would accelerate disease progression is the subject of much speculation. True resistance appears to be mediated by changes in HIV-1 gp120 that allow binding to the complex of the CCR5 receptor and bound drug.74,75
Maraviroc-resistant HIV-1 variants have been generated by serial passage in vitro.76 For one virus isolate, 2 mutations in the V3 loop, T316A and V323I, were associated with maraviroc resistance; a third V3 loop mutation, A319S, was not consistently observed. Standard drug susceptibility testing showed a decrease in the percent maximal inhibition achievable with maraviroc, without an appreciable shift in the IC50.75 This pattern has been ascribed to the noncompetitive nature of maraviroc inhibition of gp120 binding to CCR5, and it suggests that resistant viruses have developed the capacity to bind the CCR5 receptor-drug complex. Maraviroc-resistant recombinants retained sensitivity to both aplaviroc and enfuvirtide. Aplaviroc-resistant viruses also have been isolated after serial in vitro passage. All viruses retained CCR5 tropism. However, the cells used for passage had either no or low levels of CXCR4, possibly limiting the opportunity for CXCR4 variants to emerge. In contrast to maraviroc resistance, aplaviroc resistance was characterized by a rightward shift in the IC50 and no change or plateau in the percent maximal inhibition.77 Mutations that conferred decreased sensitivity to aplaviroc did not cluster in the V3 loop and were found in the C1-5, V1, and V3 regions of gp120 and within gp41, as well. In addition, the phenotypic effects of these mutations depended on the env context in which they were expressed. That is, mutations that emerge in a given env backbone do not confer resistance when engineered into env from a different viral strain.
Clonal analysis of V3 loop sequences obtained from subjects experiencing virologic failure while taking vicriviroc showed evidence of genetic selection of the V3 loop sequences, although no common pattern of mutations could be identified across samples from different subjects.78 As in the case of maraviroc resistance, the V3 loop changes that emerged with vicriviroc treatment were associated with a decrease in the percent maximal inhibition achievable. The contribution to vicriviroc susceptibility of other envelope regions outside the V3 loop remains to be determined.
Resistance to CCR5 antagonists emerges as viruses acquire the ability to use the inhibitor-bound form of CCR5 for viral entry.74,75 Decreases in the percent maximal suppression seen with the emergence of CCR5 antagonist resistance is a function of the allosteric, noncompetitive nature of small-molecule CCR5 antagonism and reflects the ability of the virus to use either inhibitor-bound CCR5 or unbound CCR5 for entry. As the virus becomes more efficient at using antagonist-bound CCR5 for entry, the height of the plateau decreases.74 Eventually, plateau height will decrease below 50%, making it impossible to calculate an IC50 for the resistant virus
| Consequences of CCR5 blockade | Top of page |
Although G-protein–coupled receptors have been targets of pharmacologic inhibition in the past, chemokine inhibitors are the first antiretroviral drugs that target host proteins. The apparent absence of significant immunologic deficits amongst individuals with naturally occurring mutations (ie, CCR5-Δ32 homozygotes) that result in a lack of functional CCR5 provides some reassurance that pharmacologic blockade of CCR5 will be relatively benign.79 Presumably, redundancy in the chemokine network allows other chemokine receptors to subsume the function of CCR5. However, pharmacologic blockade of a receptor in mature individuals may have different consequences than congenital absence of the receptor. Thus, the long-term safety of CCR5 blockade remains to be proven.
Several studies of CCR5 knockouts in mice as well as epidemiologic data from human CCR5-Δ32 homozygotes suggest this deletion may have previously unrecognized consequences. For example, although no overt pathologic changes were noted in a mouse CCR5 knockout, alterations of macrophage function and increased susceptibility to cryptococcal infections of the brain have been reported.80,81 Moreover, increased mortality from West Nile virus encephalitis was seen in CCR5–/– mice and was linked to decreased leukocyte trafficking into the brain.82 Mice that are CCR5 deficient also have an abnormal immune response to ocular infection with herpes simplex virus type 1.83 In a mouse model of T cell –mediated hepatitis, CCR5 deficiency increased mortality and liver injury.84 The lack of signaling due to the absence of CCR5 was thought to prevent downregulation of the natural killer T-cell response, resulting in fulminant hepatitis. The relevance of this finding to the cases of aplaviroc-induced hepatotoxicity is unclear.
The murine studies have been corroborated by studies in humans showing that CCR5Δ32 heterozygotes have a six-fold increased risk for severe morbidity from West Nile virus infection and a five-fold increased risk of mortality.85 The immunologic importance of functional CCR5 is underscored by several studies demonstrating improved outcomes in patients with immunologically mediated conditions such as rheumatoid arthritis and organ transplantation, among others, in CCR5Δ32 homozygotes.86,87
| Moving Forward—Anticipated Clinical Uses of CCR5 Antagonists | Top of page |
For at least the next 1 to 2 years, and perhaps into the foreseeable future, maraviroc is the only FDA-approved CCR5 inhibitor available for clinical use. As such, a discussion of its most appropriate use in 3 different segments of the patient population is warranted.
| Antiretroviral-Naive Patients | Top of page |
Evidence of increased rates of treatment failures has been reported for both vicriviroc and maraviroc. Given the impressive potency of currently recommended first-line regimens, and the theoretical concerns of adverse events (ie, infection, malignancy with vicriviroc), the use of a CCR5 antagonist in antiretroviral-naive patients is currently not recommended.
Antiretroviral-Experienced Patients—Salvage and Deep Salvage
Most of the clinical data with maraviroc have been derived from its use in this patient population. As such, the available phase 3 data support the use of maraviroc for this patient subgroup. All patients being considered for maraviroc therapy should first be screened by a tropism assay. Results demonstrating the presence of dual-mixed or X4 virus should preclude the use of maraviroc. Since achieving an undetectable viral load is the goal of therapy in all patients, maraviroc will best be used when it can be combined with at least 2, and preferably 3, other active drugs. The availability of newer-generation protease, nonnucleoside reverse transcriptase, and integrase inhibitors makes this a more realistic goal in patients for perhaps the first time in years, if ever. A recent comparison of once- and twice-daily maraviroc in the MOTIVATE studies suggested that twice-daily dosing was more effective, and the drug labeling recommends maraviroc be dosed twice daily.88 Because of drug interactions, the dose of maraviroc must be adjusted for concomitant medications as follows: 300 mg twice daily (nucleoside reverse transcriptase inhibitor [NRTI], enfuvirtide, tipranavir/ritonavir, nevirapine); 150 mg twice daily (all other ritonavir-boosted PIs); and 600 mg twice daily (efavirenz, rifampin).
Antiretroviral-Experienced Patients—Second-Line Therapy
Given that these types of patients were not included in MOTIVATE 1 or 2, it is difficult to make informed recommendations on the use of maraviroc in this population. Points arguing for its use include the greater likelihood of patients having pure R5 virus earlier in the infection and the considerable viral load reductions over 24 weeks when used in patients with more advanced disease.
With all patients, theoretical concerns over long-term drug safety should be openly discussed with patients as part of a broader discussion and their education regarding all available treatment options at the time of first failure. All patients considering R5 antagonist therapy should first be screened with a tropism assay; detection of dual/mixed or X4 virus precludes their use.
| Conclusions | Top of page |
The CCR5 antagonists are a welcome addition to the therapeutic armamentarium available for antiretroviral-experienced patients. Currently, their use in antiretroviral-naive patients should be restricted to enrollment in ongoing or planned clinical trials. The CCR5 antagonist maraviroc is FDA-approved for treatment-experienced patients with R5 virus (only), and no patient should receive maraviroc without first undergoing a tropism assay. Although the cost of this additional test is substantial, the possibility of combining maraviroc with at least 2 other active drugs to achieve undetectable viral loads may justify the added expense. Ongoing clinical monitoring will provide important follow-up data to address the theoretical safety concerns surrounding CCR5 inhibition. The approval of drugs from several new drug classes onto the market in 2007 through 2008 will herald a major step forward in the treatment of antiretroviral-experienced patients with multidrug-resistant HIV-1.
| References | Top of page |
