For those lucky enough to be in attendance, the keynote lecture delivered at the 6th Conference on Retroviruses and Opportunistic Infections (CROI) at the end of January 1999 was nothing short of a jaw-dropping experience. Dr. Beatrice Hahn of the University of Alabama at Birmingham presented the first concrete evidence of the primate origin of HIV-1, the much more prevalent of the HIV types responsible for the AIDS pandemic. The official report of her team’s findings, published in a February 1999 issue of Science, was a no less captivating example of seminal scientific research (Gao, 1999).
Since 1999, Dr. Hahn's group has been working tirelessly in the laboratory and in the chimpanzee communities of sub-Saharan Africa to illuminate the pathways leading to the emergence of HIV-1, the adaptive changes that followed, and the mechanisms underlying its pathogenicity in humans. The genetic similarities between chimpanzees and humans-they share more than 98% sequence identity across their genomes (Watanabe, 2004)-and the newly demonstrated relationship between HIV and SIV may be useful to researchers searching for key differences in virus-host interactions that may explain why and how HIV causes immune deficiency in humans, while SIV is nonpathogenic to its natural host. It is this research that continues to guide current studies evaluating the susceptibility of humans to zoonoses such as AIDS and may guide the development of new treatments and vaccines.
| The Origin of HIV and SIV | Top of page |
There are two known genetically distinct AIDS viruses: human immunodeficiency virus-1 (HIV-1) and human immunodeficiency virus-2 (HIV-2). HIV-1 is divided into three major clades, groups M, N and O; group M is the clade most widely distributed and associated with the majority of disease globally. Both HIV-1 and HIV-2 are of primate origin. The origin of HIV-2 has been established to be the sooty mangabey (Cercocebus atys), an Old World monkey of Guinea Bissau, Gabon, and Cameroon (Hirsch, 1989; Gao, 1992). The origin of HIV-1 is the central common subspecies of chimpanzee (see Figure 1).
Figure 1.Pan troglodytes: The Primate Source of HIV-1

Both HIV-1 and HIV-2 are of primate origin. The origin of HIV-2 has been established to be the sooty mangabey, an Old World monkey of Guinea Bissau, Gabon, and Cameroon. The origin of HIV-1 is the central subspecies of chimpanzee, pictured here.
Photo courtesy of Karl Ammann. Published with permission of Goldray Consulting Group.
"The reason we know this is because these primates harbor viruses that are genetically very closely related to the human viruses," Dr. Hahn said. "What most people don't know is that there are at least 38 other primate species in sub-Saharan Africa, each harboring their own version of what is called simian immunodeficiency virus, or SIV." Chimpanzees acquired SIV from two smaller primates, the greater spot-nosed monkey (Cercopithecus petaurista) and the red-capped mangabey (Cercocebus torquatus). "The chimpanzee virus is a recombinant of ancestors of these other viruses," she added. "Chimpanzees acquired their infection like humans did, by hunting and consuming naturally infected primates."
Dr. Hahn pointed out the term SIV is a misnomer. "We called it SIV because it's so closely related to HIV, which we discovered first," she said. "However, SIV is not an immunodeficiency virus-it does not cause immune deficiency, or AIDS, in its natural host."
| Primate Lentiviruses | Top of page |
HIV is an RNA virus that codes for the enzyme reverse transcriptase, which is needed to transcribe the viral RNA into a DNA copy that is capable of integrating itself into the host cell genome. Within the retrovirus family, HIV is classified as a lentivirus. It has morphologic and genetic similarities to animal lentiviruses, including those infecting cats (feline immunodeficiency virus), sheep (visna virus), goats (caprine arthritis-encephalitis virus), and non-human primates (SIV). "When we compare the sequences of these lentiviruses, phylogenetic analysis yields clusters revealing common ancestors," Dr. Hahn explained. "The human viruses cluster with SIV; they do not cluster with the lentiviruses in other animals. However, the cluster that we do see indicates that the human viruses have to be the result of cross-species transmission, meaning that they clearly came from primates."
The primate lentiviruses for which full-length genomic sequences are available fall into several major, approximately equidistant, phylogenetic lineages. "When you compare different primate lentiviruses, what you see is that the viruses cluster according to their species," Dr. Hahn explained. "When all chimp viruses cluster together, or all sooty mangabey viruses cluster together, it's easy to recognize when they cross over to a different species. In addition, what has been observed is that some closely related species have closely related viruses. This has given rise to the hypothesis that these viruses are very old. Perhaps they are as old as the species, to the point where some viruses and some hosts coevolved. And that would put a timeline of approximately a million years on some of these. It's also clear that there is cross-species transmission; not just to humans, but among non-human primates themselves."
One limitation of the research conducted thus far is the fact that most SIVs are derived from primates studied in captivity. This, Dr. Hahn argued, does not provide information concerning the prevalence, genetic diversity, and geographic distribution in the wild. "What we're left with are questions concerning the magnitude of the existing SIV reservoirs and the associated human zoonotic risk," she said.
| The Origin of HIV: Pan troglodytes | Top of page |
Chimpanzees can be divided into two species: the common chimpanzee (Pan troglodytes) and the bonobo (Pan paniscus). Pan troglodytes can be divided into four subspecies: Pan troglodytes schweinfurthii (P. t. schweinfurthii), also known as the eastern common chimpanzee; Pan troglodytes troglodytes (P. t. troglodytes), or the central common chimpanzee; Pan troglodytes verus (P. t. versus), or the western common chimpanzee; and Pan troglodytes vellerosus (P. t. vellerosus), or the Nigeria chimpanzee. The geographic locations of the four Pan troglodytes subspecies in sub-Saharan Africa are detailed in Figure 2.
Figure 2.Pan Troglodytes Subspecies
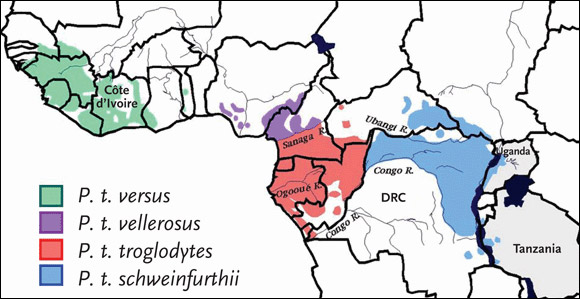
Chimpanzees can be divided into two species: the common chimpanzee (Pan troglodytes) and the bonobo (Pan paniscus). Pan troglodytes can be divided into four subspecies: Pan troglodytes schweinfurthii (P. t. schweinfurthii), also known as the eastern common chimpanzee; Pan troglodytes troglodytes (P. t. troglodytes), or the central common chimpanzee; Pan troglodytes verus (P. t. versus), or the western common chimpanzee; and Pan troglodytes vellerosus (P. t. vellerosus), or the Nigeria chimpanzee.
Source: Sharp, 2005. Journal of Virology 79(7):3892. Adapted with permission of the American Society for Microbiology.
Prior to 1999, only three wild-born Pan troglodytes tested in laboratories had been found to harbor SIV (SIVcpz). Two of the chimpanzees (GAB1 and GAB2), caught wild in Gabon, had viruses with similar genotypes. The third chimpanzee (Noah), confiscated in Antwerp after having been illegally imported from Zaire (now the Democratic Republic of Congo), had a variant of SIVcpz that differed genotypically from the SIVcpz variants found in GAB1 and GAB2 (Huet, 1990; Jansenns, 1994; Jin, 1994).
In 1999, Dr. Hahn’s group sequenced the genome of a fourth SIVcpz strain, isolated from frozen samples taken from an African-born chimpanzee (Marilyn) who died of sepsis after having given birth to stillborn twins in 1985 while housed at an American primate center (Gao, 1999). Marilyn’s sepsis and the stillbirths were not associated with immune deficiency, as SIV does not cause immune deficiency in chimpanzees. With access to equipment intended for mitochondrial DNA analysis, Dr. Hahn’s group also determined Marilyn’s subspecies, along with the subspecies of the three other SIVcpz-infected Pan troglodytes.
The analysis revealed that GAB1, GAB2, and Marilyn harbored viruses closely related to each other. All were members of the subspecies P. t. troglydytes. Noah, who harbored the genetically divergent SIVcpz strain, belonged to the subspecies P. t. schweinfurthii. Phylogenetic analyses also revealed that all HIV-1 strains known to infect humans, including HIV-1 groups M, N, and O, were closely related to just one of these SIVcpz lineages: that found in P. t. troglodytes.
According to a recent review by Drs. Paul Sharp, George Shaw, and Hahn, detailed phylogenetic analyses of HIV-1 diversity in west equatorial Africa have provided additional evidence for a P. t. troglodytes origin of HIV-1 (Sharp, 2005). The three highly divergent HIV-1 strains—groups M, N, and O—are more closely related to SIVcpz from P. t. troglodytes than from P. t. schweinfurthii. Remarkably, these three HIV-1 groups were not each other’s closest relatives but instead were interspersed with SIVcpz within the HIV-1/SIVcpz radiation (see Figure 3). This finding suggests that there were three separate introductions of SIVcpz into the human population, all from the central common subspecies of chimpanzee.
Figure 3.Evolutionary Relationships of SIVcpz and HIV-1 Strains
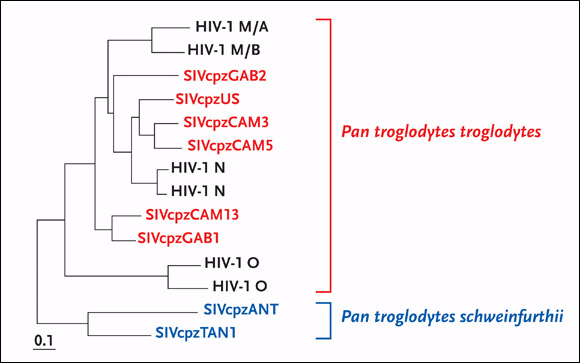
The evolutionary relationships of SIVcpz and HIV-1 strains are based on maximum-likelihood phylogenetic analyses of full-length envelope protein sequences. SIVcpz strains from P. t. troglodytes are featured in blue at the top of phylogenetic tree, whereas SIVcpz strains from P. t. schweinfurthii are featured in red at the bottom of the tree. Representative strains of HIV-1 groups M, N, and O are included for comparison.
Source: Sharp, 2005. Journal of Virology 79(7):3892. Adapted with permission of the American Society for Microbiology.
Consistent with this suggestion was the observation that HIV-1 groups N and O are largely restricted to west equatorial Africa (group N has been reported only in Cameroon), coincident with the natural range of the central chimpanzee subspecies. HIV-1 group M has been spread globally, yet the greatest extent of HIV-1 group M genetic diversity has been reported in Kinshasa, the capital of the Democratic Republic of Congo, which has led to the suggestion that the epidemic began in this geographic region. While wild chimpanzees are not found in the immediate vicinity of Kinshasa, this city of nearly 7 million people is situated on the Congo River, which likely allowed for the easy transport of SIV-infected bushmeat and of infected humans from rural to urban areas (see Figure 4).
Figure 4.Bushmeat in Pointe Noire, Democratic Republic of Congo
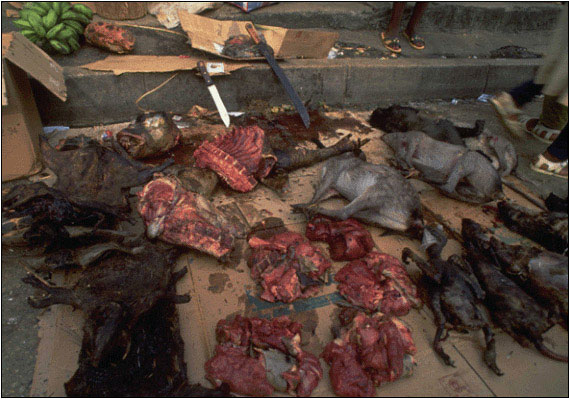
It has been theorized that SIV moved from chimpanzees and sooty mangabeys to humans—evolving into pathogenic HIV-1 and HIV-2 respectively--through exposure to primate blood, most likely as a result of the bushmeat trade. The HIV-1 group M epidemic likely began in the region of Kinshasa, Democratic Republic of Congo. Although wild chimpanzees are not found in the immediate vicinity of Kinshasa, the city is situated on the Congo River, which allowed for the easy transport of SIV-infected bushmeat and of infected humans from rural to urban areas. At this bushmeat market in Pointe Noire, a butchered chimpanzee is shown in the middle of the photograph, along with other smoked and fresh meat.
Photo courtesy of Karl Ammann. Published with permission of Goldray Consulting Group.
These results demonstrated that the SIVcpz-infected P. t. troglodytes is the natural origin of HIV-1 and is the source of at least three independent introductions of SIVcpz into the human population.
| Natural SIVcpz Reservoirs | Top of page |
Interestingly, SIV prevalence among chimpanzees in captivity is unexpectedly low. More than 2,000 wild-born chimpanzees in captivity have been tested and only eight full-length sequences from SIV-infected chimpanzees are available: six from P. t. troglodytes and two from P. t. schweinfurthii. “SIVcpz infection appears to be very rare,” Dr. Hahn commented. “After 15 years of studying this, all we have in the database are eight sequences. This definitely isn’t for lack of looking.”
The rarity of SIVcpz infection has led to a number of important questions: Do chimpanzees represent a natural SIV reservoir? If so, what are the prevalence, geographic distribution, and genetic diversity of SIVcpz in the wild? How, why, when, and where was SIVcpz transmitted to humans? Finally, were adaptive changes required to transform SIVcpz into an epidemically important human pathogen?
Answers to these questions can only be found in one place: chimpanzees’ natural habitats. However, the evaluation of chimpanzees in the wild presents many challenges. First, they are an endangered species; poaching and lost natural habitat have decimated their numbers. Second, their habitat is remote and fragmented. They live in communities consisting of five to 150 individuals in isolated forest regions. They are also reclusive animals and tend to avoid human contact, except for the rare instances when they have been habituated to human observers.
| Testing in the Wild | Top of page |
All of these factors suggested that collecting blood samples from chimpanzees was not feasible. In turn, Dr. Hahn’s group developed methods to identify SIVcpz-specific RNA and antibodies in chimpanzee urine samples and fecal matter collected from the forest. “We’re lucky because antibodies are present in both feces and urine,” Dr. Hahn explained.
In a pilot study of SIVcpz-infected captive chimpanzees, antibodies were found—using commercially available Western blot—in 63% of fecal and 100% of urine samples, suggesting that this approach was of sufficient sensitivity and specificity for field testing (Santiago, 2002; Santiago, 2003). SIV-RNA was also documented in 56% of fecal samples. While the sensitivity did not approach that of antibody testing, all of the SIVcpz-infected chimpanzees studied had at least one PCR-positive fecal sample, suggesting that SIV-RNA analysis could be used for molecular confirmation of infection, especially if multiple samples from the same individuals could be collected and tested.
| Gombe National Park | Top of page |
With the noninvasive screening assays at the ready, Dr. Hahn’s team headed to Gombe National Park in Tanzania. “It’s probably the most famous chimp colony in the world,” Dr. Hahn explained. “This is where Dr. Jane Goodall started her work over forty years ago. Gombe is wonderful because many of the chimps are habituated to the presence of human observers. They go about their business and they aren’t bothered when someone follows them and records what they do on a daily basis.”
Dr. Hahn explained how her study team collects urine from wild chimps. “Trees in the forest contain nests,” she said. “The chimps sleep in the nests, in order to avoid being attacked by leopards during the night. We would wake up in the morning before the chimps and stand under the trees with collection baskets. Basically, they do the same thing we do in the morning. We’d capture the urine in the baskets and collect the necessary sample amount. As for feces, we’d collect it off the jungle floor and preserve it for testing.”
Gombe National Park is a very small national park, approximately 675 kilometers by 70 kilometers, located on the shores of Lake Tanganyika. “It’s pretty much an island of forest in Tanzania,” Dr. Hahn explained. “Everything to the north, south, and east of Gombe National Park has been deforested, meaning that there’s no place for the chimpanzees to go. This is bad, because the chimps can’t get out and there is concern of inbreeding.”
There are three P. t. schweinfurthii communities within Gombe National Park: the northern Mitumba community, which has about 20 chimps; the central Kasekela community, which has 57 chimps; and the southern Kalande community, which has approximately 20 chimps. “Mitumba and Kasakela are habituated to humans,” Dr. Hahn explained. “Kalande is not. The Kalande chimps have not yet been habituated to tolerate the presence of human observers. However, we have been collaborating with other researchers in Gombe who have been able to collect samples for us. We’ve pretty much tested every chimp in Gombe so far.”
An initial survey of 76 of the 95 Gombe chimpanzees yielded SIVcpz prevalence estimates of 17%, 5%, and 30% for the Mitumba, Kasekela, and Kalande communities, respectively (Santiago, 2003). Infection was confirmed by urine antibody and/or fecal SIV-RNA analysis in seven of these individuals. Two additional chimpanzees exhibited urine antibody profiles suggestive of infection. More recent studies of the entire Gombe chimpanzee population with more sensitive methods identified 11 additional infections, suggesting an overall SIVcpz prevalence throughout Gombe of approximately 20%: 20%, 9%, and 50% for the Mitumba, Kasekela, and Kalande communities respectively.
Most of the infections were identified in the Kalende community. Because Kalende is not habituated, it was virtually impossible to visually link collected samples with the individual chimps. “The chimps would disappear before anyone was able to identify who did what,” Dr. Hahn explained. To identify and deduplicate the chimps, Dr. Hahn’s group conducted mitochondrial DNA testing of the samples. “We would extract the fecal DNA,” she explained. “We performed mitochondrial analysis to confirm that the sample was from a chimp. We got a haplotypes for that. We also got genomic DNA. With this, we could determine the gender of the chimp. We also did microsatellite analysis. All of this together provided us with a genotype. In turn, we could name these chimps based on what we know about them genetically.” Thus far, 8/18 (44%) individual chimps in the Kalende community have been found to harbor SIVcpz.
These field studies in Gombe were important because they revealed for the first time a prevalence of SIVcpz that begins to approximate that found in other primates naturally infected with SIVs. Moreover, with approximately half of the Kalende community members infected, it appeared that hotspots of SIVcpz infection might exist.
“These communities in Gombe are truly separate, in the sense that there is a single alpha male within each community,” Dr. Hahn explained. “All the males stay put. It’s the females that migrate, usually before they become sexually mature. This is to help avoid inbreeding. The females wander but the males stay put. The way these separate communities interact is by exchanging females, which is probably how the virus spreads. Exactly how SIVcpz is transmitted in these communities we don’t know, but it’s one of the things we’re studying.”
In summarizing her study team’s experiences in Gombe, Dr. Hahn touched upon a few valuable lessons. “This work represents the first time anyone has documented endemic infection in a wild setting,” she said. “The viruses that we identified extended the geographic range of SIVcpz to the eastern-most limits of the chimpanzee range. It provided genetic evidence for the existence of P. t. schweinfurthii lineage that did not server as the zoonotic source of HIV-1, because it’s genetically too divergent.” Moreover, she continued, this research “allowed for the field evaluation of noninvasive tests for determining prevalence of nonhabituated areas. These nonhabituated sites are ideal for long-term studies of the natural history of SIVcpz in its natural host—work that is ongoing.”
| Beyond Gombe | Top of page |
After the completion of data collection and analysis in Gombe, Dr. Hahn’s team ventured to forested areas in the vicinity of Kisangani on the Congo River, home to other communities of P. t. schweinfurthii. While Gombe National Park represents the eastern range of Pan troglodytes, the Kisangani forests are more centrally located. Here, Dr. Hahn’s group documented the presence of SIVcpz infection in two of three field sites.
Additional field surveys have been conducted (Sharp, 2005). They included the habituated communities in Kanyawara and Ngogo, both in Kibale National Park in Western Uganda; habituated chimpanzees in the Budongo Forest, in northern Uganda; habituated members of the M group in Mahale Mountain National Park in southern Tanzania; and nonhabituated chimps in the Nyungwe Forest Reserve in western Uganda. “We got multiples samples from known individuals and, surprisingly, did not find a single shred of virus,” Dr. Hahn said. “It’s possible that the virus was in these communities once, but has gone extinct.” Alternatively, she explained, the eastward spread of SIVcpz may have been influenced by biogeographical barriers, such that certain communities were never exposed to this virus.
| Prevalence of SIVcpz in P. t. troglodytes | Top of page |
Until recently, noninvasive testing of SIVcpz infection in wild-living chimpanzees from west central Africa—home to a sizeable population of P. t. troglodytes—has been limited, primarily because of a lack of established field sites with appropriate research infrastructure. Over the past few years, however, Dr. Hahn’s group, working with investigators from the University of Montpellier, have collected and analyzed samples collected from wild chimpanzees at different field sites in Cameroon.
They have confirmed the presence of P. t. vellerosus and P. t. troglodytes, using mitochondrial DNA analysis, and have used Western blot analysis to identify SIVcpz antibody-positive fecal samples. PCR analysis has also been performed to analyze fecal RNA, and microsatellite analysis has been completed for individual identification and sample enumeration. This has allowed for SIVcpz prevalence determinations at different sites and phylogenetic characterization of newly infected SIVcpz strains. However, these data have not yet been published in a peer-reviewed scientific journal and, as a result, cannot yet be reported in the pages of The PRN Notebook.
What can be discussed here is that, over the past decade, more than 100 wild-born P. t. troglodytes and 50 P. t. vellerosus chimpanzees have been screened in wildlife centers and sanctuaries (Sharp, 2005). While only a few naturally infected P. t. troglodytes were identified, molecular analysis of these viruses has yielded important insight into the evolutionary history of SIV—and HIV—in west central Africa.
First, SIVcpz infection has been identified in captive members of P. t. troglodytes but not P. t. vellerosus. While this cannot conclusively establish that P. t. vellerosus does not harbor SIVcpz, it does suggest that natural infection is rare in this subspecies. Second, SIVcpz strains from P. t. troglodytes in neighboring areas in southern Cameroon and northern Gabon were found to exhibit significant genetic diversity, suggesting longstanding infection in this area. Finally, phylogenetic analyses provided evidence of ancient recombination among members of the P. t. troglodytes SIVcpz lineage, at a time when the precursors of HIV-1 groups M, N, and O were still infecting chimpanzees. As Dr. Hahn’s group will demonstrate when its data are published, the significant clustering of HIV-1 groups M and N with SIVcpz in parts of Cameroon point to a geographic origin for these viruses.
| Conclusion | Top of page |
Even with emerging data regarding the geographical origins of HIV-1 groups M, N, and O, there are still unanswered questions regarding the natural SIV reservoirs that were the source of infection in chimpanzees. At the same time, very little is known about the biology of SIVcpz infection in chimpanzees, nor is there much data to elucidate the circumstances by which SIVcpz was transmitted to humans and gave rise to the HIV-1 epidemic. It is also unclear if SIVcpz underwent adaptive changes in order to become pathogenic and to spread as it did in the human population. If SIVcpz is nonpathogenic in chimpanzees, then elucidating the molecular basis for the differences between SIVcpz infection of apes and HIV-1 infection of humans demands attention. And if adaptive changes in SIVcpz were required, research into these changes may very well lead to a deeper understanding of HIV-1 transmission and pathogenesis, potentially guiding the ongoing search for novel prevention and treatment strategies.
| References | Top of page |
